Deconvolution1D¶
Overview¶
This benchmark is based on a 1D Deconvolution test problem from the library CUQIpy. It defines a posterior distribution for a 1D deconvolution problem, with a Gaussian likelihood and four different choices of prior distributions with configurable parameters.
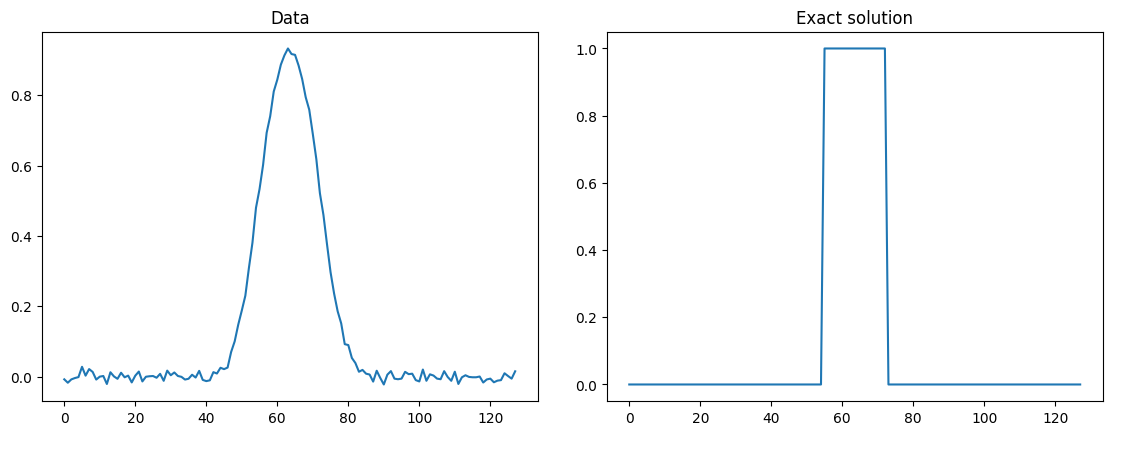
Plot of data and exact solution
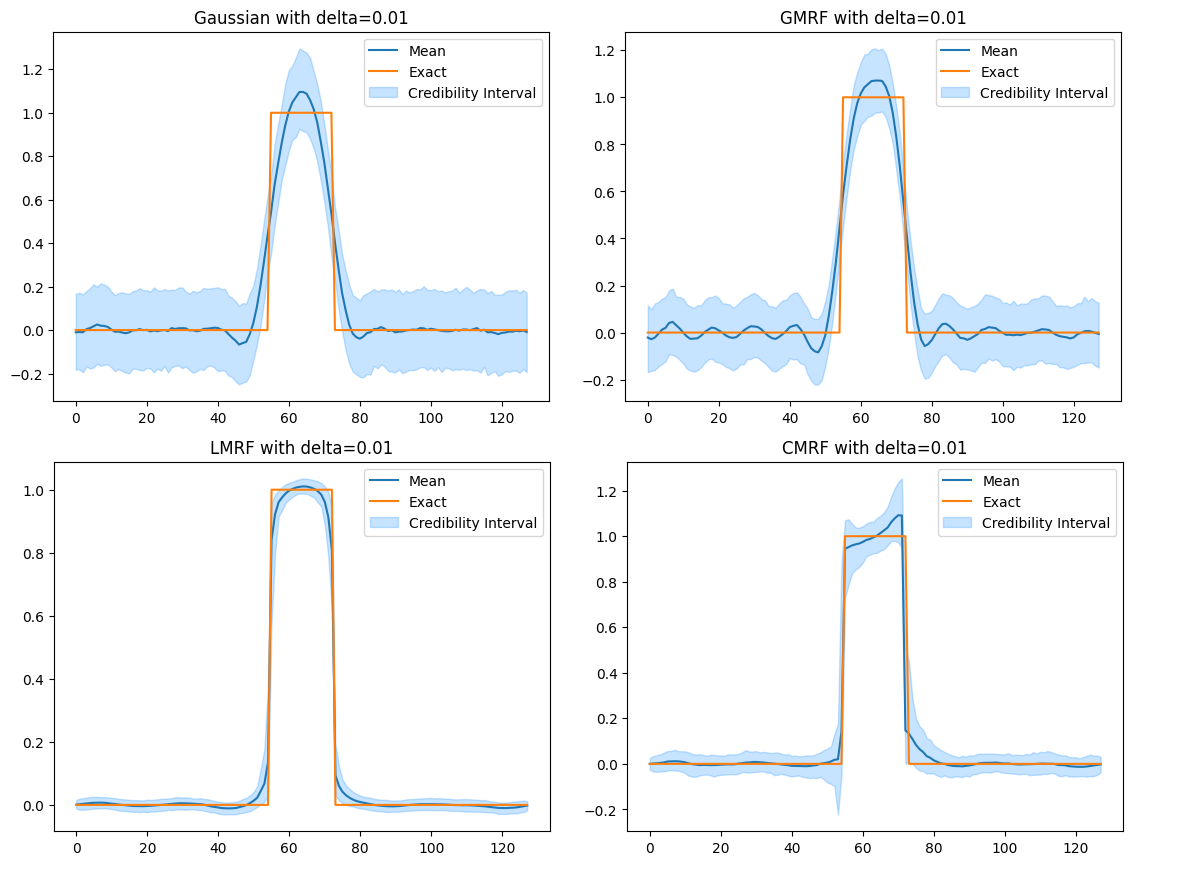
Credibility interval plots of posterior samples using different priors
Run¶
docker run -it -p 4243:4243 linusseelinger/benchmark-deconvolution-1d:latest
Properties¶
Model |
Description |
|---|---|
Deconvolution1D_Gaussian |
Posterior distribution with Gaussian prior |
Deconvolution1D_GMRF |
Posterior distribution with Gaussian Markov Random Field prior |
Deconvolution1D_LMRF |
Posterior distribution with Laplacian Markov Random Field prior |
Deconvolution1D_CMRF |
Posterior distribution with Cauchy Markov Random Field prior |
Deconvolution1D_ExactSolution |
Exact solution to the deconvolution problem |
Deconvolution1D_Gaussian¶
Mapping |
Dimensions |
Description |
|---|---|---|
input |
[128] |
Signal \(\mathbf{x}\) |
output |
[1] |
Log PDF \(\pi(\mathbf{b}\mid\mathbf{x})\) of posterior with Gaussian prior |
Feature |
Supported |
|---|---|
Evaluate |
True |
Gradient |
True |
ApplyJacobian |
False |
ApplyHessian |
False |
Config |
Type |
Default |
Description |
|---|---|---|---|
delta |
double |
0.01 |
The prior parameter \(\delta\) (see below). |
Deconvolution1D_GMRF¶
Mapping |
Dimensions |
Description |
|---|---|---|
input |
[128] |
Signal \(\mathbf{x}\) |
output |
[1] |
Log PDF \(\pi(\mathbf{b}\mid\mathbf{x})\) of posterior with Gaussian Markov Random Field prior |
Feature |
Supported |
|---|---|
Evaluate |
True |
Gradient |
True |
ApplyJacobian |
False |
ApplyHessian |
False |
Config |
Type |
Default |
Description |
|---|---|---|---|
delta |
double |
0.01 |
The prior parameter \(\delta\) (see below). |
Deconvolution1D_LMRF¶
Mapping |
Dimensions |
Description |
|---|---|---|
input |
[128] |
Signal \(\mathbf{x}\) |
output |
[1] |
Log PDF \(\pi(\mathbf{b}\mid\mathbf{x})\) of posterior Laplacian Markov Random Field prior |
Feature |
Supported |
|---|---|
Evaluate |
True |
Gradient |
False |
ApplyJacobian |
False |
ApplyHessian |
False |
Config |
Type |
Default |
Description |
|---|---|---|---|
delta |
double |
0.01 |
The prior parameter \(\delta\) (see below). |
Deconvolution1D_CMRF¶
Mapping |
Dimensions |
Description |
|---|---|---|
input |
[128] |
Signal \(\mathbf{x}\) |
output |
[1] |
Log PDF \(\pi(\mathbf{b}\mid\mathbf{x})\) of posterior with Cauchy Markov Random Field prior |
Feature |
Supported |
|---|---|
Evaluate |
True |
Gradient |
True |
ApplyJacobian |
False |
ApplyHessian |
False |
Config |
Type |
Default |
Description |
|---|---|---|---|
delta |
double |
0.01 |
The prior parameter \(\delta\) (see below). |
Deconvolution1D_ExactSolution¶
Mapping |
Dimensions |
Description |
|---|---|---|
input |
[0] |
No input to be provided. |
output |
[128] |
Returns the exact solution \(\mathbf{x}\) for the deconvolution problem. |
Feature |
Supported |
|---|---|
Evaluate |
True |
Gradient |
False |
ApplyJacobian |
False |
ApplyHessian |
False |
Config |
Type |
Default |
Description |
|---|---|---|---|
None |
Mount directories¶
Mount directory |
Purpose |
|---|---|
None |
Source code¶
Description¶
The 1D periodic deconvolution problem is defined by the inverse problem
where \(\mathbf{b}\) is an \(m\)-dimensional random vector representing the observed data, \(\mathbf{A}\) is an \(m\times n\) matrix representing the convolution operator, \(\mathbf{x}\) is an \(n\)-dimensional random vector representing the unknown signal, and \(\mathbf{e}\) is an \(m\)-dimensional random vector representing the noise.
This benchmark defines a posterior distribution over \(\mathbf{x}\) given \(\mathbf{b}\) as
where \(\pi(\mathbf{b}|\mathbf{x})\) is a likelihood function and \(\pi(\mathbf{x})\) is a prior distribution.
The noise is assumed to be Gaussian with a known noise level, and so the likelihood is defined via
where \(\mathbf{I}_m\) is the \(m\times m\) identity matrix and \(\sigma=0.01\) defines the noise level.
The prior can be configured by choosing of the following assumptions about \(\mathbf{x}\):
Gaussian: Gaussian (Normal) distribution: \(x_i \sim \mathcal{N}(0, \delta)\).GMRFGaussian Markov Random Field: \(x_i-x_{i-1} \sim \mathcal{N}(0, \delta)\).CMRFCauchy Markov Random Field: \(x_i-x_{i-1} \sim \mathcal{C}(0, \delta)\).LMRFLaplace Markov Random Field: \(x_i-x_{i-1} \sim \mathcal{L}(0, \delta)\).
where \(\mathcal{C}\) is the Cauchy distribution and \(\mathcal{L}\) is the Laplace distribution. The parameter \(\delta\) is the prior parameter and is configurable (see above).
The choice of prior is specified by providing the name to the HTTP model. In this case Deconvolution1D_Gaussian, Deconvolution1D_GMRF, Deconvolution1D_CMRF, and Deconvolution1D_LMRF, respectively. See um-bridge Clients for more details.
In addition to the HTTP models for the posterior, there is also an HTTP model for the exact solution to the problem. This model is called Deconvolution1D_ExactSolution and returns exact phantom used to generate the synthetic data when called.
In CUQIpy the benchmark is defined and sampled as follows:
import numpy as np
import cuqi
# Parameters
delta = 0.01
prior = "LMRF"
# Load data, define problem and sample posterior
TP = cuqi.testproblem.Deconvolution1D(
dim=128,
PSF="gauss",
PSF_param=5,
phantom="square",
noise_std=0.01,
noise_type="gaussian"
)
if prior == "Gaussian":
TP.prior = cuqi.distribution.Gaussian(np.zeros(128), delta)
elif prior == "GMRF":
TP.prior = cuqi.distribution.GMRF(np.zeros(128), 1/delta)
elif prior == "CMRF":
TP.prior = cuqi.distribution.CMRF(np.zeros(128), delta)
elif prior == "LMRF":
TP.prior = cuqi.distribution.LMRF(np.zeros(128), delta)
TP.sample_posterior(2000).plot_ci(exact=TP.exactSolution)